I’ve been a fan of ghostKOALA for a while, but uploading batches of 300mb files to a web interface was a pain. Enter kofamscan! KEGG published hmm profiles of each KEGG ortholog and implemented a tool to search amino acid sequences for them. There is still a web interface, but there is also an command line tool.
The documentation states that kofamscan needs to be run on linux, but I was successful running it on unix using conda.
I downloaded the databases and executables using:
wget ftp://ftp.genome.jp/pub/db/kofam/ko_list.gz # download the ko list
wget ftp://ftp.genome.jp/pub/db/kofam/profiles.tar.gz # download the hmm profiles
wget ftp://ftp.genome.jp/pub/tools/kofamscan/kofamscan.tar.gz # download kofamscan tool
wget ftp://ftp.genome.jp/pub/tools/kofamscan/README.md # download README
And then I unzipped and untarred the relevant files:
gunzip ko_list.gz
tar xf profiles.tar.gz
tar xf kofamscan.tar.gz
Next, I made a conda environment using miniconda:
conda create -n kofamscan hmmer parallel
conda activate kofamscan
conda install -c conda-forge ruby
I copied the template config file and added the relative paths of my newly downloaded kofamscan databases.
config.yml:
# Path to your KO-HMM database
# A database can be a .hmm file, a .hal file or a directory in which
# .hmm files are. Omit the extension if it is .hal or .hmm file
profile: ./profiles
# Path to the KO list file
ko_list: ko_list
# Path to an executable file of hmmsearch
# You do not have to set this if it is in your $PATH
# hmmsearch: /usr/local/bin/hmmsearch
# Path to an executable file of GNU parallel
# You do not have to set this if it is in your $PATH
# parallel: /usr/local/bin/parallel
# Number of hmmsearch processes to be run parallelly
cpu: 8
Lastly, I ran kofamscan:
./exec_annotation -f mapper -o norp88.txt norp88.faa
This gives an output file that looks like this:
NORP88_1
NORP88_2
NORP88_3 K01869
NORP88_4
NORP88_5
NORP88_6 K07082
NORP88_7
NORP88_8 K07448
NORP88_9
NORP88_10 K07507
This output is handy because it can be fed into tools like KEGG Mapper, or KEGGDecoder (binderized repo here).
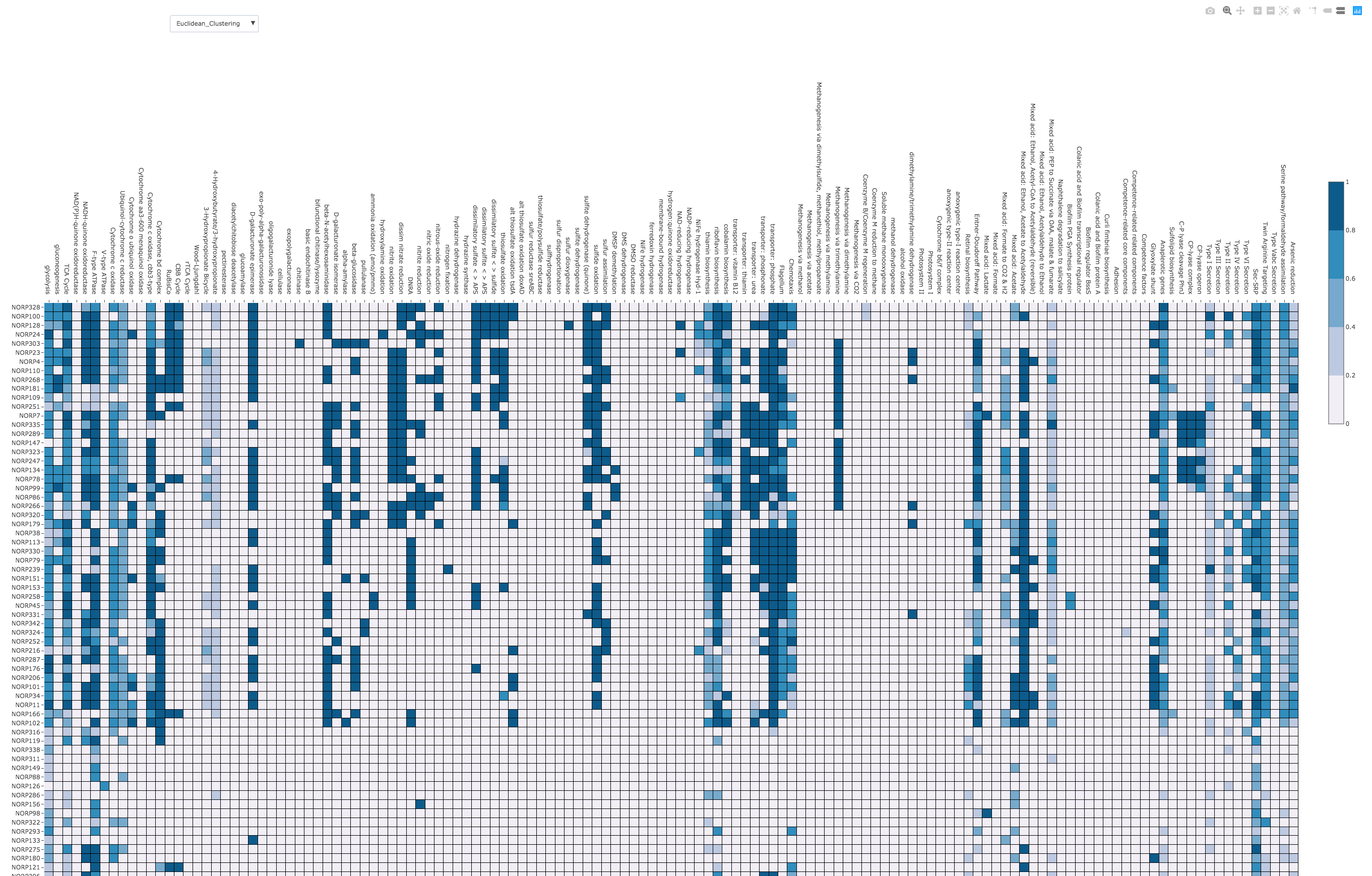
If you would prefer more information in the output, you can run kofamscan like this:
./exec_annotation -o norp88.txt norp88.faa
Which will give you an output that looks like this:
# gene name KO thrshld score E-value KO definition
#-------------------- ------ ------- ------ --------- ---------------------
NORP88_1 K08365 144.47 110.1 3.1e-33 MerR family transcriptional regulator, mercuric resistance operon regulatory protein
NORP88_1 K19591 142.77 100.3 4.3e-30 MerR family transcriptional regulator, copper efflux regulator
NORP88_1 K13638 155.43 92.3 1.1e-27 MerR family transcriptional regulator, Zn(II)-responsive regulator of zntA
NORP88_1 K19592 232.93 81.3 2.8e-24 MerR family transcriptional regulator, gold-responsive activator of gol and ges genes
NORP88_1 K13639 94.60 71.5 2.3e-21 MerR family transcriptional regulator, redox-sensitive transcriptional activator SoxR
NORP88_1 K21902 104.73 57.9 5.2e-17 MerR family transcriptional regulator, repressor of the yfmOP operon
NORP88_1 K03713 95.73 51.6 4.1e-15 MerR family transcriptional regulator, glutamine synthetase repressor
NORP88_1 K11923 90.23 49.8 1.6e-14 MerR family transcriptional regulator, copper efflux regulator
NORP88_1 K21745 122.63 49.2 2.4e-14 MerR family transcriptional regulator, aldehyde-responsive regulator
NORP88_1 K21744 271.80 47.3 8e-14 MerR family transcriptional regulator, thiopeptide resistance regulator
NORP88_1 K19057 92.77 44.3 6.9e-13 MerR family transcriptional regulator, mercuric resistance operon regulatory protein
NORP88_1 K13640 93.23 39.6 1.7e-11 MerR family transcriptional regulator, heat shock protein HspR
NORP88_1 K19575 165.97 30.4 1.4e-08 MerR family transcriptional regulator, activator of bmr gene
NORP88_1 K22491 147.63 30.3 1.2e-08 MerR family transcriptional regulator, light-induced transcriptional regulator
NORP88_1 K21743 509.60 26.6 1.5e-07 MerR family transcriptional regulator, multidrug-efflux activator
NORP88_1 K19432 68.37 13.2 0.003 anti-repressor of SlrR
NORP88_1 K21089 347.10 11.7 0.0047 MerR family transcriptional regulator, activator of the csg genes
NORP88_2 K19340 299.70 103.5 5.2e-31 Cu-processing system ATP-binding protein
NORP88_2 K01990 225.27 101.3 3.3e-30 ABC-2 type transport system ATP-binding protein
NORP88_2 K11050 351.23 85.7 1.3e-25 multidrug/hemolysin transport system ATP-binding protein
NORP88_2 K09695 352.50 85.1 2.1e-25 lipooligosaccharide transport system ATP-binding protein
NORP88_2 K05643 1692.60 82.1 9.4e-25 ATP-binding cassette, subfamily A (ABC1), member 3
NORP88_2 K20459 365.53 77.8 3.7e-23 lantibiotic transport system ATP-binding protein
NORP88_2 K02056 573.30 75.8 1e-22 simple sugar transport system ATP-binding protein [EC:3.6.3.17]
NORP88_2 K09817 238.00 75.1 2.4e-22 zinc transport system ATP-binding protein [EC:7.2.2.-]
NORP88_2 K10441 630.83 74.6 2.6e-22 ribose transport system ATP-binding protein [EC:3.6.3.17]